In-vitro, Bioinformatics and Computational Biophysical Analyses of Antifungal Phytochemicals from Acalypha wilkesiana Variants Targeting Malassezia Lipase
DOI:
https://doi.org/10.26538/tjdr/v2i9.3Keywords:
Seborrheic dermatitis, Malassezia globosa, Acalypha wikesiana, Lipase inhibition, Molecular dockingAbstract
Purpose: Seborrheic dermatitis (SD) is a common chronic inflammatory skin disorder, often associated with the activity of lipid-hydrolyzing enzymes secreted by Malassezia globosa. Conventional antifungals often show limited efficacy and adverse effects, while the mechanisms underlying traditional remedies like Acalypha wilkesiana remain underexplored.
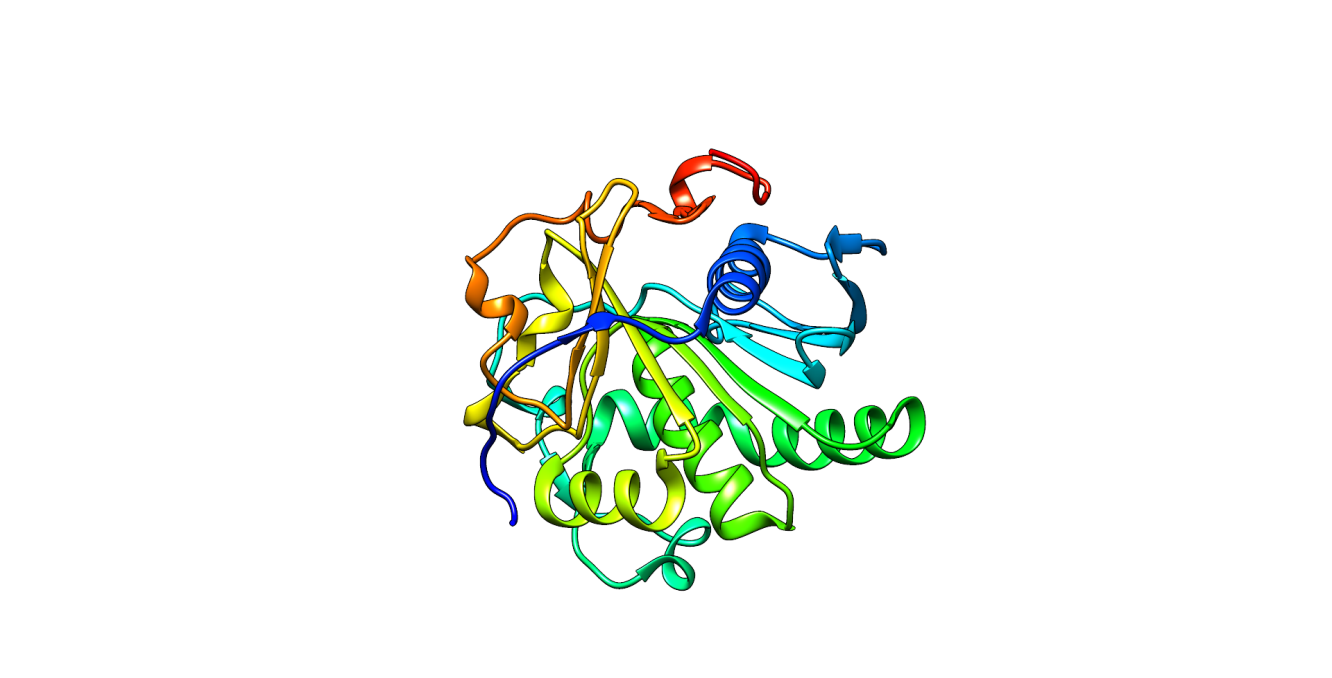
Methods: This study evaluated the antifungal potential of phytochemicals derived from the red and green variants of A. wikesiana, targeting M. globosa lipase (PDB ID: 3UUE). Crude ethanol extracts were analyzed via gas chromatography–mass spectrometry (GC-MS), and preliminary in-vitro antifungal activity was assessed against M. globosa. Identified compounds were subjected to molecular docking and molecular dynamics (MD) simulations to assess binding affinity and stability. ADMET profiling and density functional theory (DFT) calculations were employed to predict pharmacokinetics, toxicity, and electronic reactivity.
Results: The red A. wikesiana extract demonstrated strong antifungal activity, producing a 60 mm inhibition zone, while the green variant was inactive. Molecular docking identified aspidospermidin-17-ol, 1-acetyl-19,21-epoxy-15,16-dimethoxy- (ASP) as the top-performing compound with a binding affinity of –6.8 kcal/mol, outperforming ketoconazole and the native ligand. MD simulations confirmed the structural stability of the ASP–lipase complex, exhibiting minimal RMSD fluctuations. ADMET predictions indicated low dermal and systemic toxicity, and DFT analysis confirmed favorable electronic properties.
Conclusion: The findings highlight ASP from red A. wikesiana as a potent and safe inhibitor of M. globosa lipase, supporting its potential development as a novel, plant-based topical antifungal agent for the management of seborrheic dermatitis.
Downloads
References
1. Dall’Oglio F, Nasca MR, Gergino C, Micali G. An overview of diagnosis and management of seborrheic dermatitis. Clin Cosmet Investig Dermatol. 2020;15:1537-1848.
2. Schoch JJ, Monin RL, Satcher KG, Harris J, Triplett E, New J. The infantile cutaneous microbiome: a review. Pediatr Dermatol. 2019;36(5):574-580.
3. Araya M, Kutthanan K, Jiamton S. Clinical characteristics and quality of life of seborrheic dermatitis patients in a tropical country. Indian J Dermatol. 2015;60(5):5-19.
4. Rhimi W, Theeler B, Boekhout T, Otranto D, Catarchia C. Malassezia spp.: yeasts of emerging concern in fungemia. Front Cell Infect Microbiol. 2020;10:370.
5. Park M, Park S, Jung WH. Skin commensal fungus Malassezia and its lipases. J Microbiol Biotechnol. 2021;31(5):637-644.
6. Sparber F, De Gregorio C, Steckholzer S, Cilata M, Salluto F, Gut-Landmann SL. The skin commensal yeast Malassezia triggers a type 17 response that coordinates antifungal immunity and exacerbates skin inflammation. Cell Host Microbe. 2019;25:389-403.
7. Park M, Park S, Jung WH. Skin commensal fungus Malassezia and its lipases. J Microbiol Biotechnol. 2021;31(5):637-644.
8. Adalsteinsson JA, Kaushik S, Muzumdar S, Guttman-Yassky E, Ungar J. An update on the microbiology, immunology and genetics of seborrheic dermatitis. Exp Dermatol. 2020;29:481-489.
9. Mustarichie R, Rostinawati T, Pitaloka DAE, Saptarini NM, Iskandar Y. Herbal therapy for the treatment of seborrheic dermatitis. Clin Cosmet Investig Dermatol. 2022;15:2391-2405.
10. Sulaiman AN, James AE. Phytochemical constituents and antifungal activity of Acalypha wilkesiana leaves on Candida albicans. J Sci. 2023;7(3):209-214.
11. Madziga HA, Chiromo M, Sanni S, Sandabe UK, Sodipo OA. Effects of aqueous leaf extract of Acalypha wilkesiana on some serum biochemical profiles of mice in acute toxicity studies. Sahel J Vet Sci. 2020;17(3):1-5.
12. Sherifat KO, Itohan AM, Adeola SO, Adeola KM, Aderemi OL. Antifungal activity of Acalypha wilkesiana: a preliminary study of fungal isolates of clinical significance. Afr J Infect Dis. 2022;16(1):21-30.
13. Isaac AO, Tolupe AC. Phytochemical and antifungal screening of Acalypha wilkesiana Mull Arg (Euphorbiaceae) leaf extract in cream formulations. Niger J Pharm Appl Sci Res. 2022;5(1):36-41.
14. Katibi OS, Aboh MI, Sclawu OA, Kola-Mustapha A, Olatunyi LA. Antifungal activity of Acalypha wilkesiana: a preliminary study of fungal isolates of clinical significance. Afr J Infect Dis. 2022;16(1):21-30.
15. Uloma UUN, Ilomuanya MO, Salem AI, Mercy A, Oludolapo K, Chukwuemeka AP, Oluwakayinsola S. Evaluation of the antimicrobial effectiveness of topical gels containing Acalypha wilkesiana. Trop J Nat Prod Res. 2021;5(8):1470-1477.
16. Romald PN, Kindo AJ, Mahalakshmi V, Umadevi U. Epidemiological pattern of Malassezia, its phenotypic identification and antifungal susceptibility profile to azoles by broth microdilution method. Indian J Med Microbiol. 2020;38(3-4):351-356.
17. Ikezu UM, Duru CE, Ikpa CB, Ibe FC. In silico discovery of dual-targeting molecules from Acalypha wilkesiana against hypertension and hyperglycemia. Discov Chem. 2025;2:86.
18. Zaputa A, Acros SR. A comparative study of McFarland turbidity standards and the Densimat photometer to determine bacterial cell density. Curr Microbiol. 2015;70:907-909.
19. Enenebeaku UE, Duru CE, Mgbemena IC, Ukwandu NCD, Nwigwe HC, Enenebeaku CK, Okotcha EN. Phytochemical evaluation and molecular docking of bioactive compounds from the roots of Dictyandra arborescens (Welw.) against Plasmodium berghei protein targets. Trop J Nat Prod Res. 2021;5(2):370-381.
20. Duru IA, Duru CE. Molecular docking of compounds in the essential oil of Ocimum gratissimum leaf against PIM-1 kinase of Escherichia coli. Eur J Adv Chem Res. 2020;1(6):1-4.
21. Ivanova L, Tammiku-Taul J, García-Sosa AT, Sidorova Y, Saarma M, Karelson M. Molecular dynamics simulations of the interactions between glial cell line-derived neurotrophic factor family receptor GFRα1 and small-molecule ligands. ACS Omega. 2018;3(9):11407-11414.
22. Pires DEV, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic properties using graph-based signatures. J Med Chem. 2015;58(9):4066-4072.
23. Swanson K, Walther P, Leitz J, Mukherjee S, Wu JC, Shivnaraine RV, Zou J. ADMET-AI: a machine learning ADMET platform for evaluation of large-scale chemical libraries. bioRxiv. 2023; Dec 28.
24. Duan Y, Yang X, Zeng X, Wang W, Deng Y, Cao D. Enhancing molecular property prediction through task-oriented transfer learning: integrating universal structural insights and domain-specific knowledge. J Med Chem. 2024;67(11):9575-9586.
25. Duru CE, Chidiebere CW. Computer-assisted discovery of tyrosinase inhibitors from turmeric and clove: an in silico study on natural skin-whitening agents and their potential toxicity. S Afr J Bot. 2024;175:669-683.
26. Sherifat KO, Itohan AM, Adeola SO, Adeola KM, Aderemi OL. Antifungal activity of Acalypha wilkesiana: a preliminary study of fungal isolates of clinical significance. Afr J Infect Dis. 2021;16(1):21-30.
27. Nwofor CN, Duru CE, Onyenwe NE. Computational identification of small molecules in Mitracarpus scaber ethanolic leaf extract with fungal keratinase inhibitory potentials. J Nat Pestic Res. 2022;2:100010.
28. Akanbi AI, Izevbigie EV, Sherif AO, Dlamini NH, Fadele LO, Oyawaluja BO. Molecular docking, ADME and SAR analysis of 383 phytochemicals in the quest for lead antidiabetic inhibitors targeting α-amylase and α-glucosidase enzymes. Trop J Drug Res. 2025;2(1):6-13.
29. Wang K, Cheng L, Li W, Jiang H, Zhang X, Liu S, Huang Y, Qiang M, Dong T, Li Y, Wang J, Feng S, Li H. Susceptibilities of Malassezia strains from pityriasis versicolor, Malassezia folliculitis and seborrheic dermatitis to antifungal drugs. Heliyon. 2020;6(6):e04203.
30. Gopu C, Chirumamilla P, Daravath SB, Vankudoth S, Taduri S. GC-MS analysis of bioactive compounds in the plant parts of methanolic extracts of Momordica cymbalaria Fenzl. J Med Plants Stud. 2021;9(3):209-218.
31. Naz S, Alam S, Ahmed W, Khan SM, Qayyum A, Sabir M, Naz A, Iqbal A, Bibi Y, Nisa S, Khalifa AS, Gharib AF, El Askary A. Therapeutic potential of selected medicinal plant extracts against multidrug-resistant Salmonella enterica serovar Typhi. Saudi J Biol Sci. 2022;29(2):941-954.
32. Abdullah A, Biswas P, Sahabuddin M, Mubasharah A, Khan DA, Hossain A, Roy T, Rafi NMR, Dey D, Hasan MN, Bibi S, Moustafa M, Shati A, Hassan H, Garg R. Molecular dynamics simulation and pharmacoinformatic integrated analysis of bioactive phytochemicals from Azadirachta indica (Neem) to treat diabetes mellitus. J Chem. 2023;4170703.
33. Kumari A, Kumar R, Sulabh G, Singh P, Kumar J, Singh VK, Ojha KK. In silico ADMET, molecular docking and molecular simulation-based study of glabridin’s natural and semisynthetic derivatives as potential tyrosinase inhibitors. Adv Tradit Med. 2023;23:733-751.
34. Chen CP, Chen CC, Huang CW, Chang YC. Evaluating molecular properties involved in transport of small molecules in stratum corneum: a quantitative structure–activity relationship for skin permeability. Molecules. 2018;23(4):911.
35. Matin MM, Kumer A, Chandro A, Akash S, Chakma U. Rhamnopyranoside pivaloyl esters as black and white fungus inhibitors: molecular docking, dynamics and ADMET analysis. Orbital Electron J Chem. 2024;16:50-61.
36. Duru IA, Duru CE. Molecular modeling and density functional theory calculation of the coordination behaviour of 4,5-dichloroimidazole with Cu(II) ion. Sci Afr. 2020;9:1-7.
Downloads
Published
Issue
Section
License

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.




